The coefficient of inbreeding (COI) is a concept that originated in the field of genetics and was applied to dog breeding to quantify the degree of inbreeding within a particular pedigree or a breed. While it’s difficult to attribute the invention of the coefficient of inbreeding to a single individual, it has been used by geneticists and breeders for many decades.
One notable early proponent of using coefficients of inbreeding in animal breeding, including dogs, was Sewall Wright, an American evolutionary geneticist who made significant contributions to the understanding of population genetics and evolutionary theory in the mid-20th century. He used some of Pearl’s papers as a basis. Wright developed mathematical models to study the effects of inbreeding and genetic drift in populations, which laid the groundwork for later applications of coefficients of inbreeding in animal breeding programs.
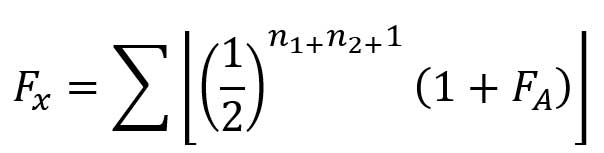
*Wright’s equation for calculating the Coefficient of Inbreeding, 1922. The equation may vary according to available data and the subject of analysis, also minding Ancestor Loss Coefficient, Mean Kinship Coefficient, or Relationship Coefficient
“If an individual is inbred, his sire and dam are connected in the pedigree by lines of descent from a common ancestor or ancestors. The coefficient of inbreeding is obtained by a summation of coefficients for every line by which the parents are connected, each line tracing back from the sire to a common ancestor and thence forward to the dam, and passing through no individual more than once. The same ancestor may of course be involved in more than one line.”
Wright, 1921
Nowadays, measuring the coefficient of inbreeding is essential for responsible dog breeders as it helps maintain genetic diversity, reduce the risk of hereditary diseases, preserve breed characteristics, improve long-term health and welfare, and adhere to ethical breeding standards.
Theoretically, COI percentages can range from 0% to 100%, following the concept of “lower is better”.
Example:
Mixed-breed dogs might have a coefficient of inbreeding around 5% if they come from unrelated breeds and individuals. The average COI for purebred dogs, representing a breed with an average to good genetic diversity, is supposed to be around 20%. The average COI for purebred dogs from a breed with poor genetic diversity tends to be around 40%. It is not recommended the COI for rare breeds to exceed 10%. According to the experts is also recommended that an individual’s COI should not exceed 6% counted from the 5-generation pedigree. Some of the highest COIs known are in the Norwegian Lundehund (over 80%) and the Doberman Pinscher (45%-60%). The COI in close inbreeding, let’s take a dog produced from a father-to-daughter mating, is 25%. Мating of first cousins produces a COI of 6.25% with the presumption of unrelated ancestors. And so on.
The optimal COI percentage can vary between breeds and even within different lines or populations of the same breed.
How can COI be measured?
Based on pedigrees
This method involves examining the dog’s pedigree to identify common ancestors shared by both parents, as these common ancestors contribute to the level of inbreeding within the pedigree.
Once common ancestors are identified, the next step is to calculate the probability of inheriting identical alleles from them. This involves determining the number of shared alleles between the ancestors and the individual dog.
The COI represents the probability that both alleles at any given locus in the dog’s genome are identical by descent, meaning they are inherited from the same ancestor.
A higher COI value suggests a higher level of inbreeding, while a lower COI value indicates a more genetically diverse background.
It’s important to note that pedigree-based calculation of COI relies on accurate and complete pedigree information, including as many generations, as possible. The accuracy of calculated COI is dependent on the number of generations. Results from 4 generations may differ from the results from 10 generations. Incomplete pedigrees or unknown ancestry can drastically affect the accuracy of COI calculations. Most COI calculators assume that the original ancestors in the pedigree are unrelated, and pedigree-based methods may not account for complex relationships or genetic contributions from individuals outside the recorded pedigree.
Additionally, because of the principle of segregation, two individuals with identical expected COIs from a pedigree may have different levels of inbreeding. It depends on the inheritance of chromosomal segments by each individual.
Based on markers
Also known as genetic marker-based COI estimation, it involves using sets of polymorphic markers to assess the genetic relatedness between individuals in a population. This method relies on molecular genetic techniques to analyze the DNA sequences or variations within an individual’s genome.
Specific genetic markers are being chosen for analysis. These markers can include single nucleotide polymorphisms, microsatellites, or other types of DNA variations that are informative for assessing genetic relatedness.
DNA samples are collected from the individuals being analyzed. This typically involves collecting a small tissue sample, such as a blood or saliva sample, from each individual.
The DNA samples are then genotyped to determine the alleles present at the selected genetic markers. Genotyping can be performed using various techniques, such as polymerase chain reaction followed by DNA sequencing or fragment analysis.
Once genotyping is complete, the genetic data for each individual are used to calculate pairwise genetic distances or genetic similarity coefficients between individuals. These measures quantify the degree of genetic relatedness between individuals based on their marker genotypes.
The COI estimation is based on the assumption that individuals with more similar marker genotypes are more closely related and therefore have a higher COI.
COI calculation based on markers offers several advantages over pedigree-based methods, including the ability to assess genetic relatedness directly from genetic data, even in the absence of complete pedigree information. Additionally, marker-based methods can provide more precise estimates of COI and are less affected by incomplete pedigrees or unknown ancestry.
However, it has some withdraws. The precise levels of genetic variation across loci are influenced by the selection of markers, with much of the genome remaining unlinked to any specific marker. Consequently, many regions of homozygosity are not detected by estimators. This limitation renders marker-based estimators ineffective in distinguishing between individuals with comparable coefficients of inbreeding (COIs).
Genome-wide coefficient of inbreeding
refers to a measure of the level of inbreeding across an individual’s entire genome, rather than focusing on specific genetic markers or regions. This concept takes advantage of advancements in genomic technologies that allow for the examination of genetic variation across an individual’s entire set of DNA. Here’s how genome-wide COI works:- Genomic data, typically in the form of DNA sequencing or genotyping data, are collected from the individual of interest. This data provides information about the genetic variation present throughout the individual's genome.
- Genomic analysis is used to identify regions of the genome where the individual is homozygous, meaning they have inherited identical alleles from both parents. Homozygous regions are indicative of recent common ancestry and are a key indicator of inbreeding.
- The proportion of the genome that consists of homozygous regions is used to calculate the genome-wide COI. This measure quantifies the extent of inbreeding across the individual's entire genome, providing a comprehensive assessment of genetic relatedness and inbreeding.
- The resulting genome-wide COI value indicates the overall level of inbreeding within the individual. Higher genome-wide COI values suggest a higher degree of recent inbreeding, while lower values indicate greater genetic diversity.
Genome-wide COI analysis offers several advantages over traditional pedigree-based or marker-based methods:
- It provides a more comprehensive assessment of inbreeding and genetic relatedness.
- It allows for the detection of smaller regions of homozygosity, providing higher resolution insights into recent common ancestry.
- It is less susceptible to biases introduced by incomplete pedigrees or marker selection, as it considers genetic variation across the entire genome.
Overall, genome-wide COI analysis is a powerful tool for assessing inbreeding and genetic relatedness, particularly in populations where traditional methods may be limited by incomplete pedigree information or genetic marker availability.



